Conformational Dynamics in the CLC Channel/Transporter Family
Core Facility: Membrane Protein Expression/Purification – Synthetic Antigen Binder (SAB) Generation and Crystallography
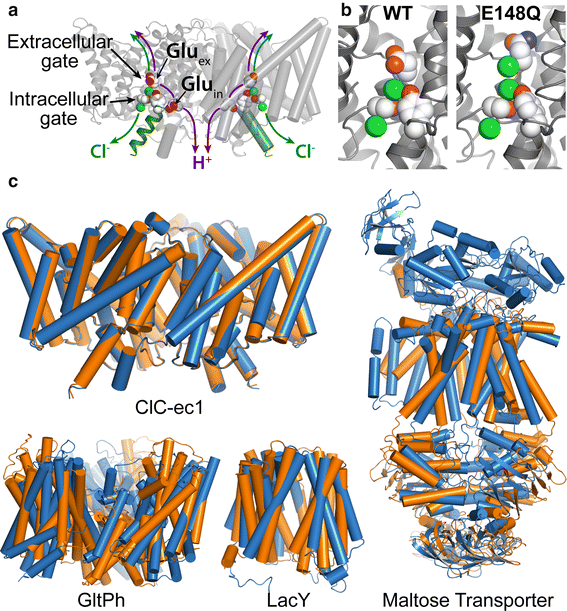
CLC proteins couple the transport of two chloride ions to the antiport of one proton in each functional cycle. CLC proteins couple the transport of two chloride ions to the antiport of one proton in each functional cycle. A complete pathway for proton transfer requires a fully connected hydrogen-bonded wire within the hydrophobic lumen of ClC. Using extended molecular dynamics simulations, mutagenesis and functional studies, the mechanism has been characterized and the pathways involved in the formation of such proton wires within the protein have been identified.
Project Description
The CLC family of chloride channels and transporters is necessary for proper neuronal, cardiovascular, and epithelial function. One of the important aspects of this family of transport proteins is that minute changes in their amino acid sequence can result in a shift in their operation mode from a channel to a transporter. Studying the structural dynamics of CLCs can therefore provide fundamental information on the nature of structural and dynamical differences between passive channels and active transporters.
The X-ray crystallographic structures of CLC proteins provide an invaluable structural foundation for understanding molecular mechanisms in this family. However, these static pictures cannot convey the structural dynamics and conformational changes necessary to carry out function. Despite crystallization under a wide variety of conditions, only one basic conformation has ever been detected. Therefore, in order to gain insight into the CLC mechanism, alternative strategies to assess conformational changes and protein dynamics are essential.
This project will address the multiple structural conformations that underlie the dual function of ClC- proteins as both channels and coupled transporters. Using a combination of NMR (solution and solid-state) and molecular dynamics simulations, the multiple conformations that support closely-coupled, stoichiometric ion transport will be accessed by binding and unbinding its two ligands, (Cl- and H+). Additional efforts will be made to use conformation-specific ligands to “lock” CLC proteins in order to study these conformations by crystallography and NMR.


