13C NMR detects conformational change in the 100-kD membrane transporter ClC-ec1
By Sherwin J. Abraham, Ricky C. Cheng, Thomas A. Chew, Chandra M. Khantwal, Corey W. Liu, Shimei Gong, Robert K. Nakamoto and Merritt Maduke.
Published in Journal of Biomolecular NMR, 2015 Jan 29. [Epub ahead of print] PMID: 25631353. PMCID: 4398623. Link to publication page.
Project: Conformational Dynamics in the CLC Channel/Transporter Family | Core Facility: Membrane Protein Expression and Purification
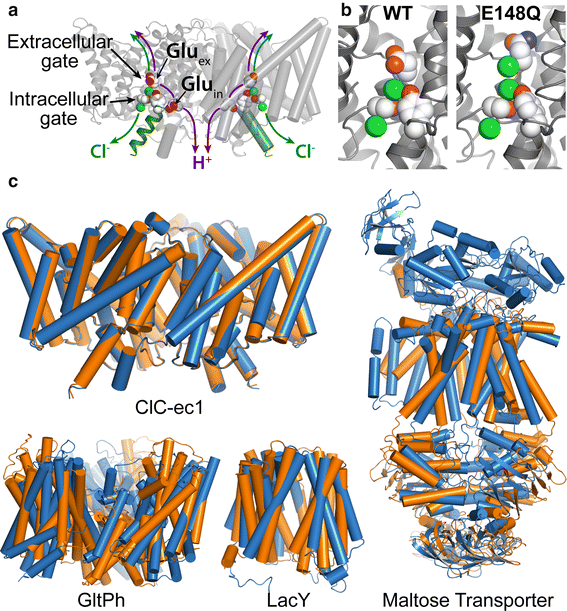
Figure 2. CLC structure and conformational change compared to conformational change detected in other membrane transporters. a X-ray crystallographic structure of ClC-ec1 (pdb ID: 1OTS). Each identical subunit catalyzes the exchange of 2 Cl- for 1 H+. The H+ pathway bifurcates from the Cl− pathway, passing along Gluex and Gluin. Gluex is also a “gate” that occludes Cl- from the extracellular solution. An intracellular Cl- gate is formed by a Ser/Tyr pair (shown in spacefill), the latter residing on Helix R (dark green), a subject of study here. b Structural comparison of WT ClC-ec1 with the Gluex → Gln mutant (E148Q). Close-up of the Cl−-binding region in WT (left) and E148Q (right). In E148Q, the glutamine side chain is flipped up out of the anion-binding site and replaced by a Cl− ion (pdb ID: 1OTU). c Structural comparison of active transporters crystallized in different conformational states. Top left: Backbone overlay of ClC-ec1 WT (orange) and E148Q (blue) structures, 0.6 Å RMSD, illustrates the lack of global conformational change in the putative outward-facing conformational state. Bottom left: GltPh, a glutamate transporter homolog (Focke et al. 2013) in outward-facing (orange) and inward-facing (blue) states (pdb 4OYE and 4P19) (Verdon et al. 2014). Bottom middle: LacY, a member of the Major Facilitator Superfamily of transporters (Yan 2013) in occluded (orange) and inward-facing (blue) conformational states (pdb 4OAA and 2V8N) (Guan et al. 2007; Kumar et al. 2014). Right: Maltose transporter, a member of the ATP-binding cassette (ABC) transporter superfamily (Chen 2013) in inward-facing (orange) and occluded (blue) states (pdb 3FH6 and 2R6G) (Khare et al. 2009; Oldham et al. 2007)
Abstract
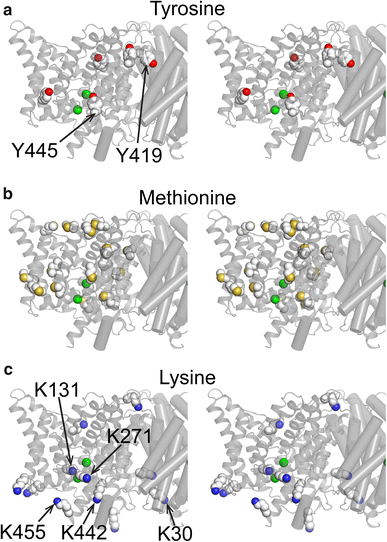
CLC transporters catalyze the exchange of Cl− for H+ across cellular membranes. To do so, they must couple Cl− and H+ binding and unbinding to protein conformational change. However, the sole conformational changes distinguished crystallographically are small movements of a glutamate side chain that locally gates the ion-transport pathways. Therefore, our understanding of whether and how global protein dynamics contribute to the exchange mechanism has been severely limited. To overcome the limitations of crystallography, we used solution-state 13C-methyl NMR with labels on methionine, lysine, and engineered cysteine residues to investigate substrate (H+) dependent conformational change outside the restraints of crystallization. We show that methyl labels in several regions report H+-dependent spectral changes. We identify one of these regions as Helix R, a helix that extends from the center of the protein, where it forms the part of the inner gate to the Cl−-permeation pathway, to the extracellular solution. The H+-dependent spectral change does not occur when a label is positioned just beyond Helix R, on the unstructured C-terminus of the protein. Together, the results suggest that H+ binding is mechanistically coupled to closing of the intracellular access-pathway for Cl−.

![Figure 3. 1H–13C HSQC spectra of 13C-Met labeled ClC-ec1. a Wildtype ClC-ec1 (homodimer, 100 kDa), methionine methyl region. b Monomeric ClC-ec1 (I201W/I422W, 50 kDa). c [H+]-dependence of 1H-13C HSQC spectra: blue, pH 7.5; orange, pH 5.0; black, reversal to pH 7.5. The asterisk and dagger indicate the most clearly resolved crosspeaks that exhibit H+-dependency. d Partially deuterated (~80 %) monomeric ClCe-c1 with prolonged data collection. e Size exclusion chromatograms of ClC-ec1. Top WT ClC-ec1; Middle monomeric ClC-ec1 (mClC-ec1) (I201W/I422W); Bottom Δ6 mClC-ec1 (mutating six Met residues as follows: M38L, M65I, M228V, M276T, M330V, and M332L) yields a mix of dimer and monomer populations](/site-media/images/publications/2015/abraham2015-figure3.gif)
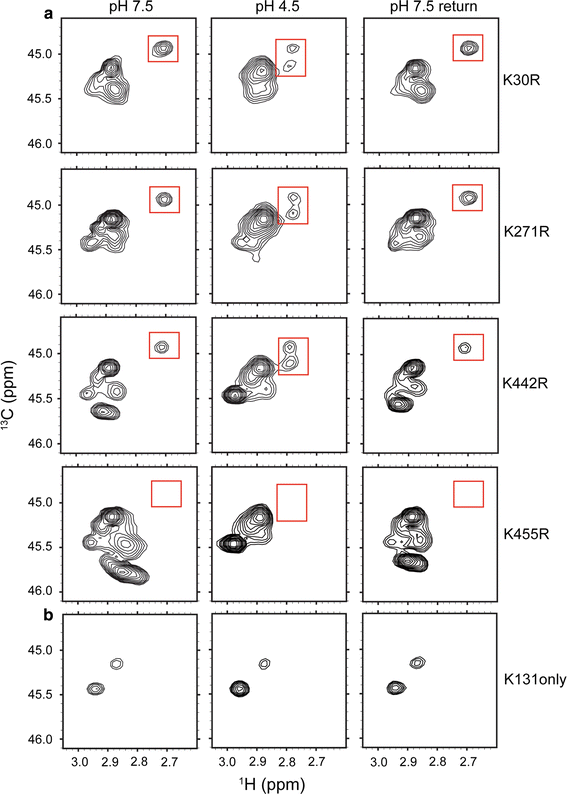
![Figure 4. 13C-Lys ClC-ec1. a 1H–13C HSQC spectra of methylated ClC-ec1 at pH 7.5 (left), pH 4.5 (middle) and reversed to pH 7.5 (right). The spectral window is zoomed in on the 13C methyl signals arising from the Lys side chains (with the 13C signal from the methylated N-terminus not shown). Changes in [H+] induce spectral shifts, the most notable being the peak splitting and shift of the peak at δH = 2.7 ppm (highlighted by the red boxes) at low pH. b pH titration of methylated ClC-ec1. Left: Overlaid 1H-13C HSQC spectra at pH 7.5, 6.5, 6.0, 5.5, 5.0, 4.5, and 4.0, as indicated in the figure. Right: 1H chemical shifts for the peak of interest as a function of pH. The solid curve is a fit to a single-site H+-binding isotherm, with an apparent pKa of 5.6. c 1H–13C HSQC spectra of methylated channel-like mutant at pH 7.5 (left), pH 4.5 (middle) and reversed to pH 7.5 (right). The spectrum of the channel-like mutant exhibits little [H+] sensitivity, and the peak at δH = 2.70 ppm and δC = 44.9 ppm is notably missing at both high and low pH](/site-media/images/publications/2015/abraham2015-figure4.gif)
![Figure 6. Functional characterization of reductively methylated ClC-ec1. a Schematic outline of the Cl− flux assay. ClC-ec1 is reconstituted into liposomes at high [KCl], and then the extravesicular solution is exchanged for a low-[KCl] solution (replacing Cl− with the impermeant isethionate, “X−”). Bulk Cl− efflux through ClC-ec1 is initiated by valinomycin (a K+ ionophore) and CCCP (a H+ ionophore) to dissipate the electrical potential. Extravesicular [Cl−] is monitored using a Ag/AgCl electrode. b Representative raw data traces for WT ClC-ec1 (red) and control liposomes (containing no protein) (black dashes). At the end of each experiment, the detergent Triton X-100 was added to lyse the liposomes and release all of the Cl−. c Summary data for reductively methylated (RM) proteins studied here. The relative turnover rates (normalized to WT unlabeled ClC-ec1) were calculated from the initial slope (Δ[Cl−]/Δt) after addition of Vln/CCCP, and averages ± SEM (n = 3–6) are reported](/site-media/images/publications/2015/abraham2015-figure6.gif)

