Rapid parameterization of small molecules using the Force Field Toolkit
By Christopher G. Mayne, Jan Saam, Klaus Schulten, Emad Tajkhorshid, and James C. Gumbart.
Published in Journal of Computational Chemistry, 34(32):2757-2770 on December 15, 2013. PMID: 24000174. PMCID: PMC3874408. Link to publication page.
Core Facility: Computational Modeling
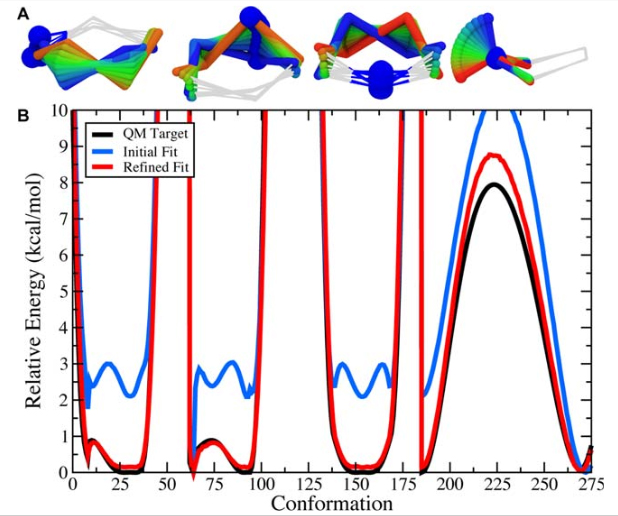
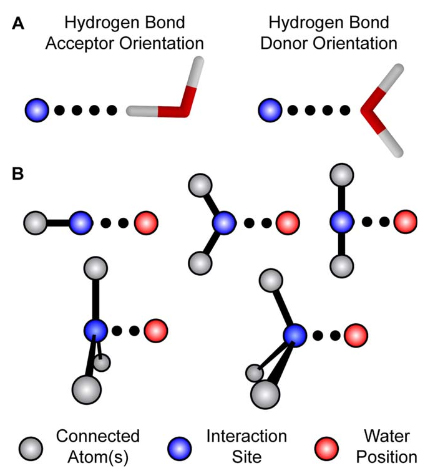
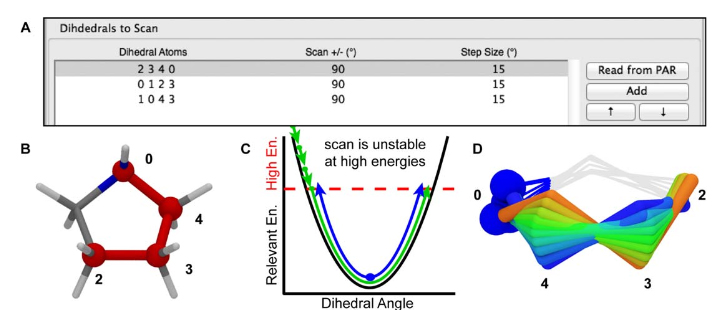
Figure 9. A) The scanned torsions for pyrrolidine were loaded into VMD and colored by iteration (blue to red) to provide a structural context for assessing the dihedral PESs. B) The cyclic structure of pyrrolidine yields a complex dihedral scan profile, in which much of the fine detail lies below 1 kcal/mol; however, an accessible barrier of 8 kcal/mol exists for the N-inversion. The refined MM parameters yield a PES (red) that reproduces the QM PES (black) with excellent agreement.
Abstract
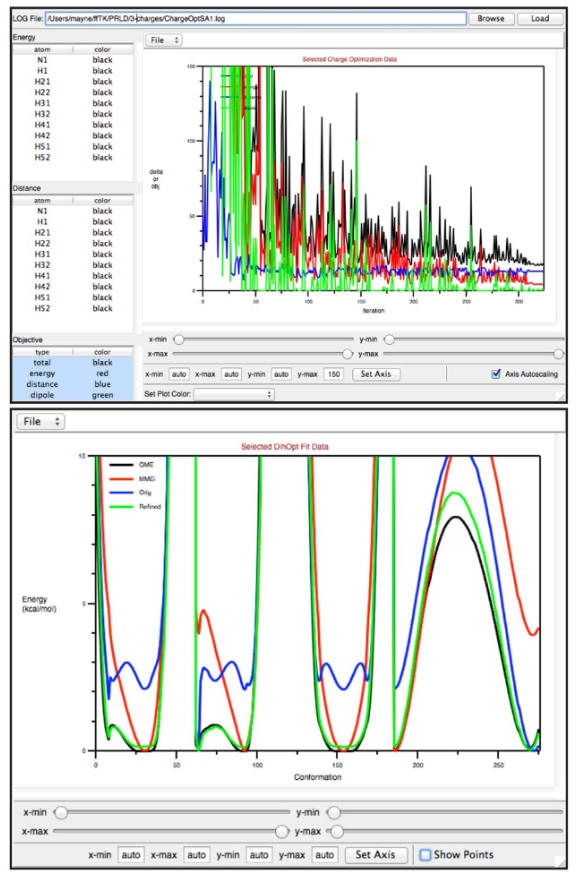
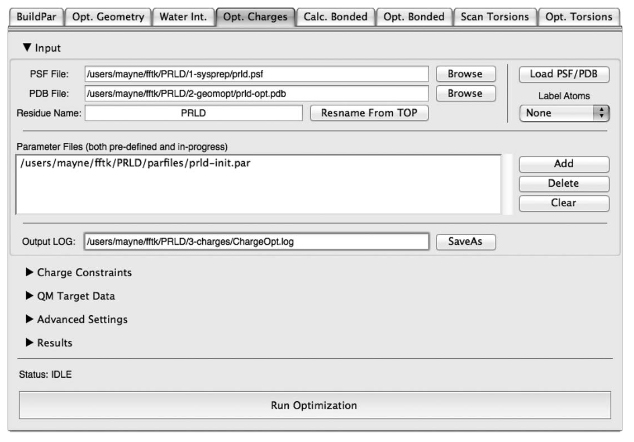
The inability to rapidly generate accurate and robust parameters for novel chemical matter continues to severely limit the application of molecular dynamics simulations to many biological systems of interest, especially in fields such as drug discovery. Although the release of generalized versions of common classical force fields, for example, General Amber Force Field and CHARMM General Force Field, have posited guidelines for parameterization of small molecules, many technical challenges remain that have hampered their wide-scale extension. The Force Field Toolkit (ffTK), described herein, minimizes common barriers to ligand parameterization through algorithm and method development, automation of tedious and error-prone tasks, and graphical user interface design. Distributed as a VMD plugin, ffTK facilitates the traversal of a clear and organized workflow resulting in a complete set of CHARMM-compatible parameters. A variety of tools are provided to generate quantum mechanical target data, setup multidimensional optimization routines, and analyze parameter performance. Parameters developed for a small test set of molecules using ffTK were comparable to existing CGenFF parameters in their ability to reproduce experimentally measured values for pure-solvent properties (<15% error from experiment) and free energy of solvation (±0.5 kcal/mol from experiment).
This publication was the Cover Article for the Journal of Computation Chemistry, Volume 34, Issue 32: