Protonation of glutamate 208 induces the release of agmatine in an outward-facing conformation of an arginine/agmatine antiporter
By Elia Zomot and Ivet Bahar
Published in The Journal of biological chemistry 286(22): 19693-701 on June 3, 2011.
PMID: 21487006. PMCID: PMC3103348. Link to Pubmed page.
Core Facility: Computational Modeling
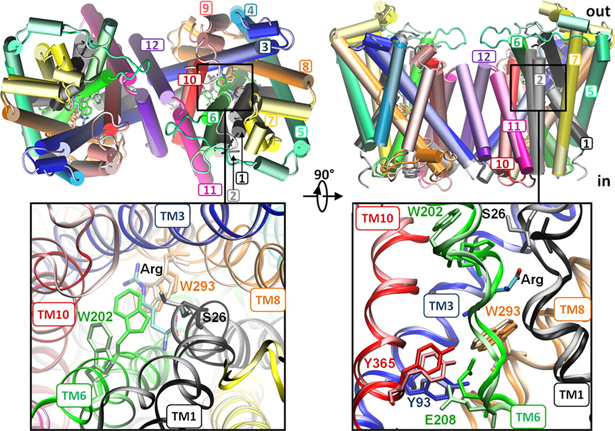
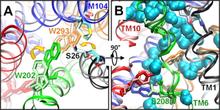
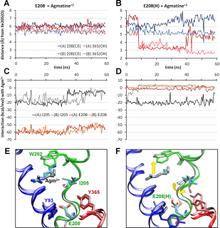
Figure 1. Open/apo and occluded/substrate-bound conformations of AdiC dimer in the outward-facing state. The apo and substrate-bound conformations (lighter and darker colors, respectively) at the beginning of the equilibration phase are superimposed, as viewed from the EC side (left) and from the side (right). Helices overlap in most cases, except for TM1, TM2, TM6, and TM10. We note in particular the large reorientation of the EC portion of TM6 in the upper left panel (see the proximal light and dark green helices on the left subunit). Structural alignment was performed on both subunits (upper panels) or on subunit A (lower panels) for more precision at the binding site. Residues controlling the EC (Trp-202, Ser-26), middle (Trp-293), and IC (Tyr-93, Tyr-365, and Glu-208) gates of the binding site (lower panels) are explicitly shown and colored according to their respective TM helices.
Abstract
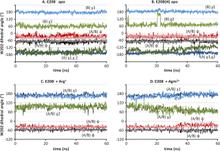
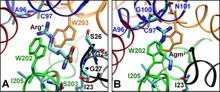
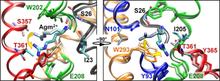
Virulent enteric pathogens have developed several systems that maintain intracellular pH to survive extreme acidic conditions. One such mechanism is the exchange of arginine (Arg(+)) from the extracellular region with its intracellular decarboxylated form, agmatine (Agm(2+)). The net result of this process is the export of a virtual proton from the cytoplasm per antiport cycle. Crystal structures of the arginine/agmatine antiporter from Escherichia coli, AdiC, have been recently resolved in both the apo and Arg(+)-bound outward-facing conformations, which permit us to assess for the first time the time-resolved mechanisms of interactions that enable the specific antiporter functionality of AdiC. Using data from ∼1 μs of molecular dynamics simulations, we show that the protonation of Glu-208 selectively causes the dissociation and release of Agm(2+), but not Arg(+), to the cell exterior. The impact of Glu-208 protonation is transmitted to the substrate binding pocket via the reorientation of Ile-205 carbonyl group at the irregular portion of transmembrane (TM) helix 6. This effect, which takes place only in the subunits where Agm(2+) is released, invites attention to the functional role of the unwound portion of TM helices (TM6 Trp-202-Glu-208 in AdiC) in facilitating substrate translocation, reminiscent of the behavior observed in structurally similar Na(+)-coupled transporters.