Visualizing Functional Motions of Membrane Transporters with Molecular Dynamics Simulations
By Saher A. Shaikh, Jing Li, Giray Enkavi, Po-Chao Wen, Zhijian Huang, and Emad Tajkhorshid.
Published in Biochemistry 52(4):569-87, on January 29, 2012. PMID: 23298176. PMCID: PMC3560430. Link to publication page.
Core Facility: Computational Modeling
Abstract
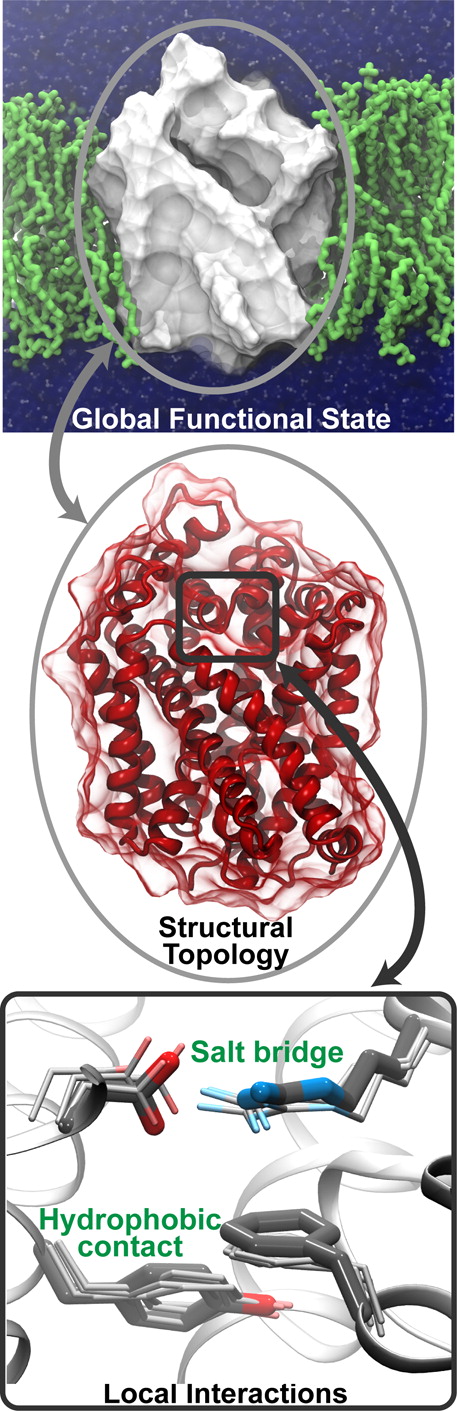
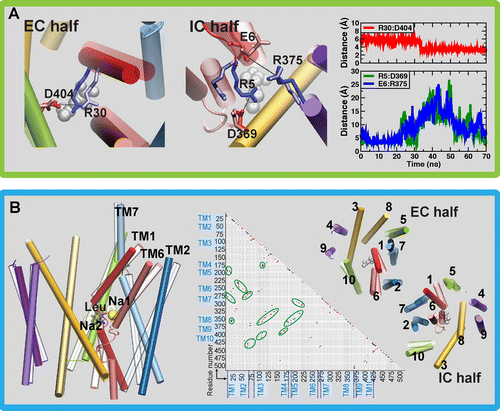
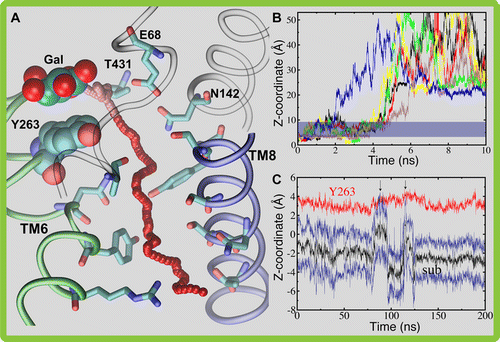
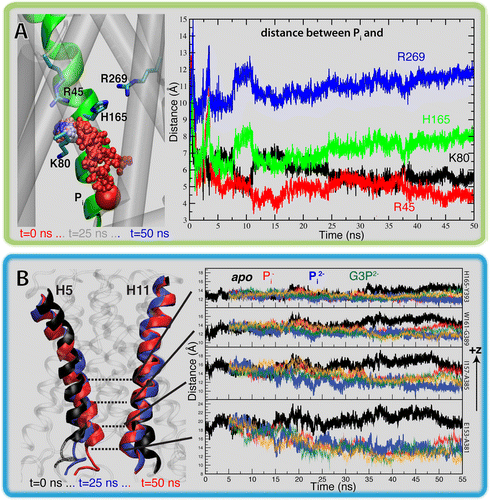
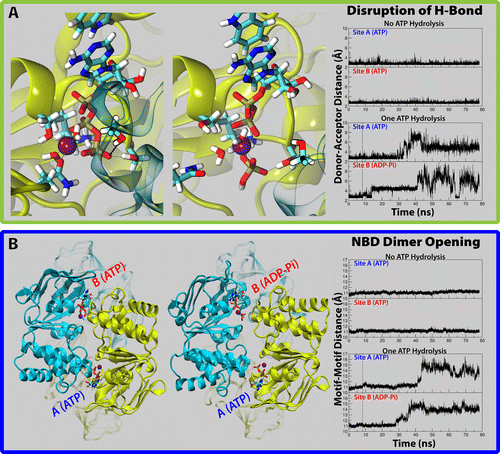
Computational modeling and molecular simulation techniques have become an integral part of modern molecular research. Various areas of molecular sciences continue to benefit from, indeed rely on, the unparalleled spatial and temporal resolutions offered by these technologies, to provide a more complete picture of the molecular problems at hand. Because of the continuous development of more efficient algorithms harvesting ever-expanding computational resources, and the emergence of more advanced and novel theories and methodologies, the scope of computational studies has expanded significantly over the past decade, now including much larger molecular systems and far more complex molecular phenomena. Among the various computer modeling techniques, the application of molecular dynamics (MD) simulation and related techniques has particularly drawn attention in biomolecular research, because of the ability of the method to describe the dynamical nature of the molecular systems and thereby to provide a more realistic representation, which is often needed for understanding fundamental molecular properties. The method has proven to be remarkably successful in capturing molecular events and structural transitions highly relevant to the function and/or physicochemical properties of biomolecular systems. Herein, after a brief introduction to the method of MD, we use a number of membrane transport proteins studied in our laboratory as examples to showcase the scope and applicability of the method and its power in characterizing molecular motions of various magnitudes and time scales that are involved in the function of this important class of membrane proteins.