Conformational Dynamics of a Glutamate Transporter Homologue
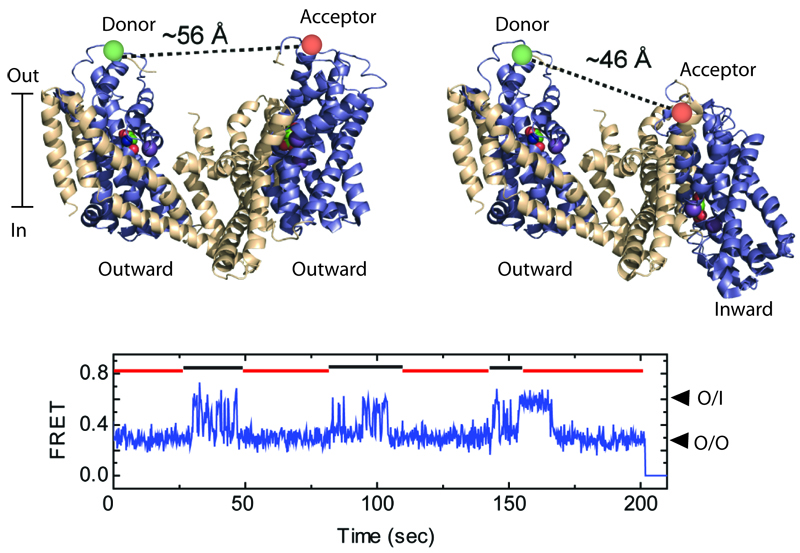
Shown are two GltPh protomers (top), labeled with the donor and acceptor fluorescent dyes, when both are in the outward-facing orientations (left), and when one of them is in an inward-facing orientation (right). The relative motions of the protomers are detected by smFRET (bottom). Such recordings reveal that protomers alternate between periods of rapid transitions and periods of quiescence, marked by black and red lines, respectively.
Project Description
Glutamate transporters reside at glutamatergic synapses in the plasma membranes of neurons and glial cells and pump the neurotransmitter from the synaptic cleft into the cytoplasm against nearly a million-fold concentration gradient. Crystallographic studies on a bacterial homologue, GltPh, have suggested that this process entails large-scale trans-membrane movements of the peripheral transport domains within the scaffold provided by three centrally located trimerization domains. Substrate and co-transported sodium ions bind to the transport domains in the outward-facing conformation, in which the binding sites are accessible from the extracellular solution. Following binding, the transport domains move over 15 Å across the lipid membranes. Next, the substrate and ions are released into the cytoplasm, and the unloaded transport domains return into the outward-facing conformation.
Trans-membrane movements of the transport domains, either when loaded with their cargo or empty, are key events of the transport cycle. This project aims to investigate the dynamics of these processes by combining single-molecule FRET (sm-FRET) imaging methods, computation and crystallography. smFRET approach allows us to visualize the domain movements in real time and has revealed a surprising dynamic complexity. Specifically, our data suggest that each transport domain resides in a stable outward- or inward-facing state for extended periods of time until it transitions into a distinct dynamic state, in which it rapidly inter-converts between the outward- and inward-facing orientations. We aim to better understand the underlying kinetic mechanism and to probe the nature of the dynamic intermediates, their functional significance and structure. We further aim to investigate the extent of coupling between the subunits in the trimer and the regulatory effects of the environmental factors, such trans-membrane electric potentials and the physico-chemical properties of the lipid bilayers.


