Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter
By Travis J. Barnard, James Gumbart, Janine H. Peterson, Nicholas Noinaj, Nicole C. Easley, Nathalie Dautin, Adam J. Kuszak, Emad Tajkhorshid, Harris D. Bernstein, and Susan K. Buchanan.
Published in Journal of Molecular Biology 415(1): 128-42 on January 6, 2012.
PMID: 22094314. PMCID: PMC3230255. Link to Pubmed page.
Core Facility: Computational Modeling
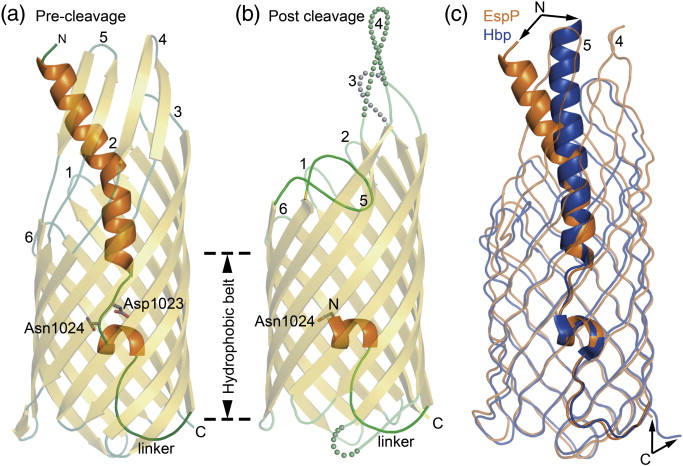
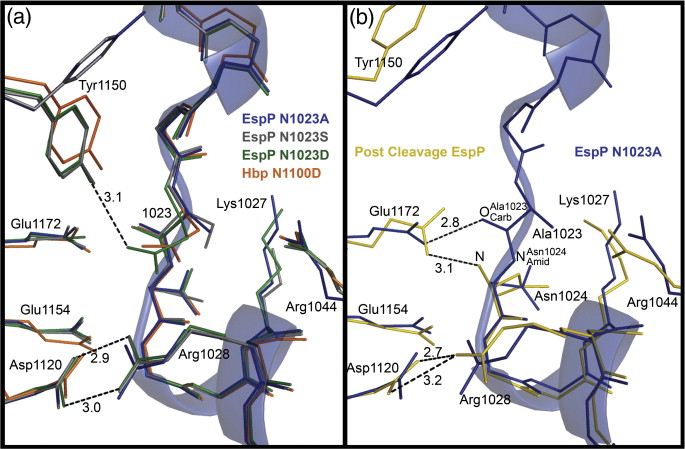
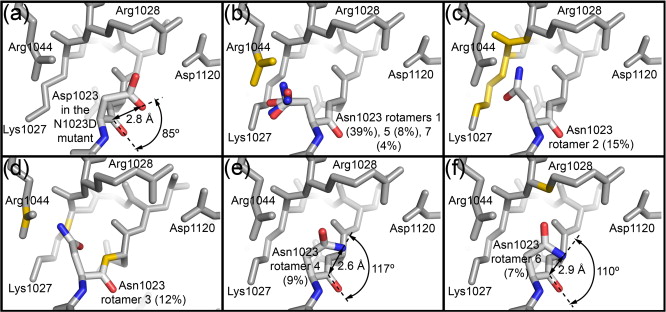
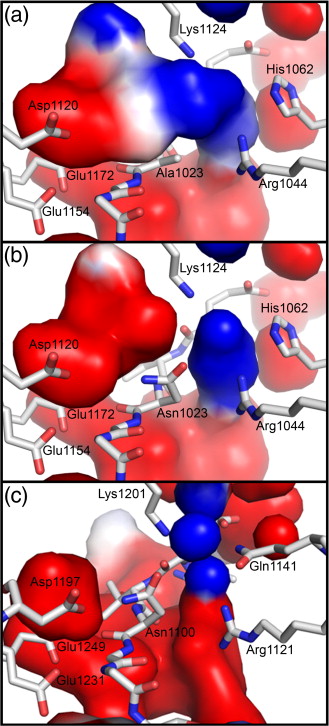
Figure 1. EspP and Hbp structures. (a) Precleavage structure of the EspP N1023D mutant. The side chains of Asp1023 and Asn1024 are shown. (b) Postcleavage structure of EspP. The N-terminal residue Asn1024 is shown. Disordered loops are depicted as spheres. (a and b) β-Strands, yellow; α-helices, orange; loops, green. The locations of the hydrophobic belt, N-terminus and C-terminus, extracellular loops 1 through 6, and linker loops are labeled. (c) Structural alignment of precleavage EspP (orange) and precleavage Hbp (blue). For EspP, loops 4 and 5 are labeled. These loops are disordered in the Hbp structure. This figure was created using the PyMOL Molecular Graphics System (Schrödinger LLC).
Abstract
Autotransporters are secreted proteins produced by pathogenic Gram-negative bacteria. They consist of a membrane-embedded β-domain and an extracellular passenger domain that is sometimes cleaved and released from the cell surface. We solved the structures of three noncleavable mutants of the autotransporter EspP to examine how it promotes asparagine cyclization to cleave its passenger. We found that cyclization is facilitated by multiple factors. The active-site asparagine is sterically constrained to conformations favorable for cyclization, while electrostatic interactions correctly orient the carboxamide group for nucleophilic attack. During molecular dynamics simulations, water molecules were observed to enter the active site and to form hydrogen bonds favorable for increasing the nucleophilicity of the active-site asparagine. When the activated asparagine attacks its main-chain carbonyl carbon, the resulting oxyanion is stabilized by a protonated glutamate. Upon cleavage, this proton could be transferred to the leaving amine group, helping overcome a significant energy barrier. Together, these findings provide insight into factors important for asparagine cyclization, a mechanism broadly used for protein cleavage.