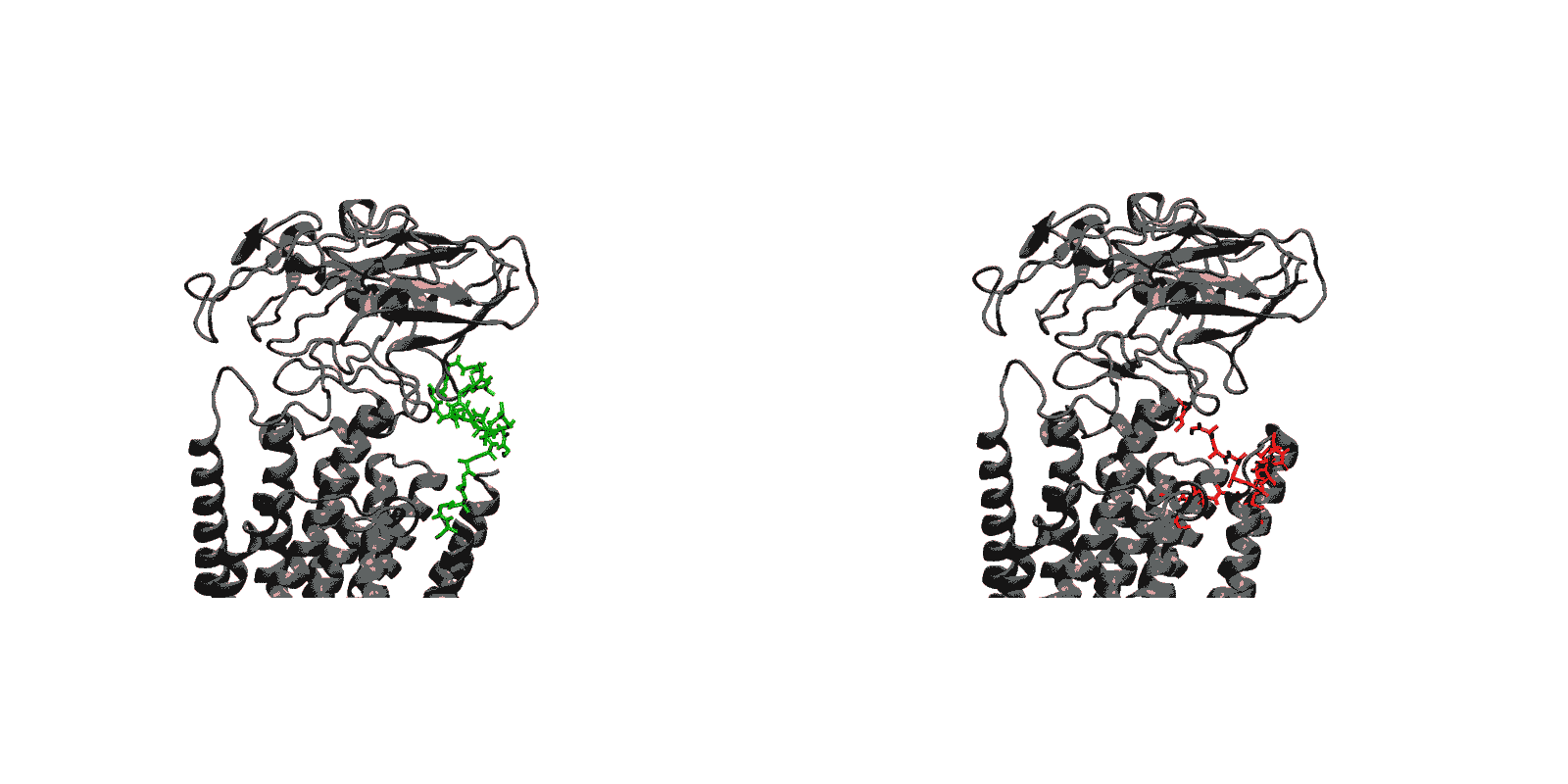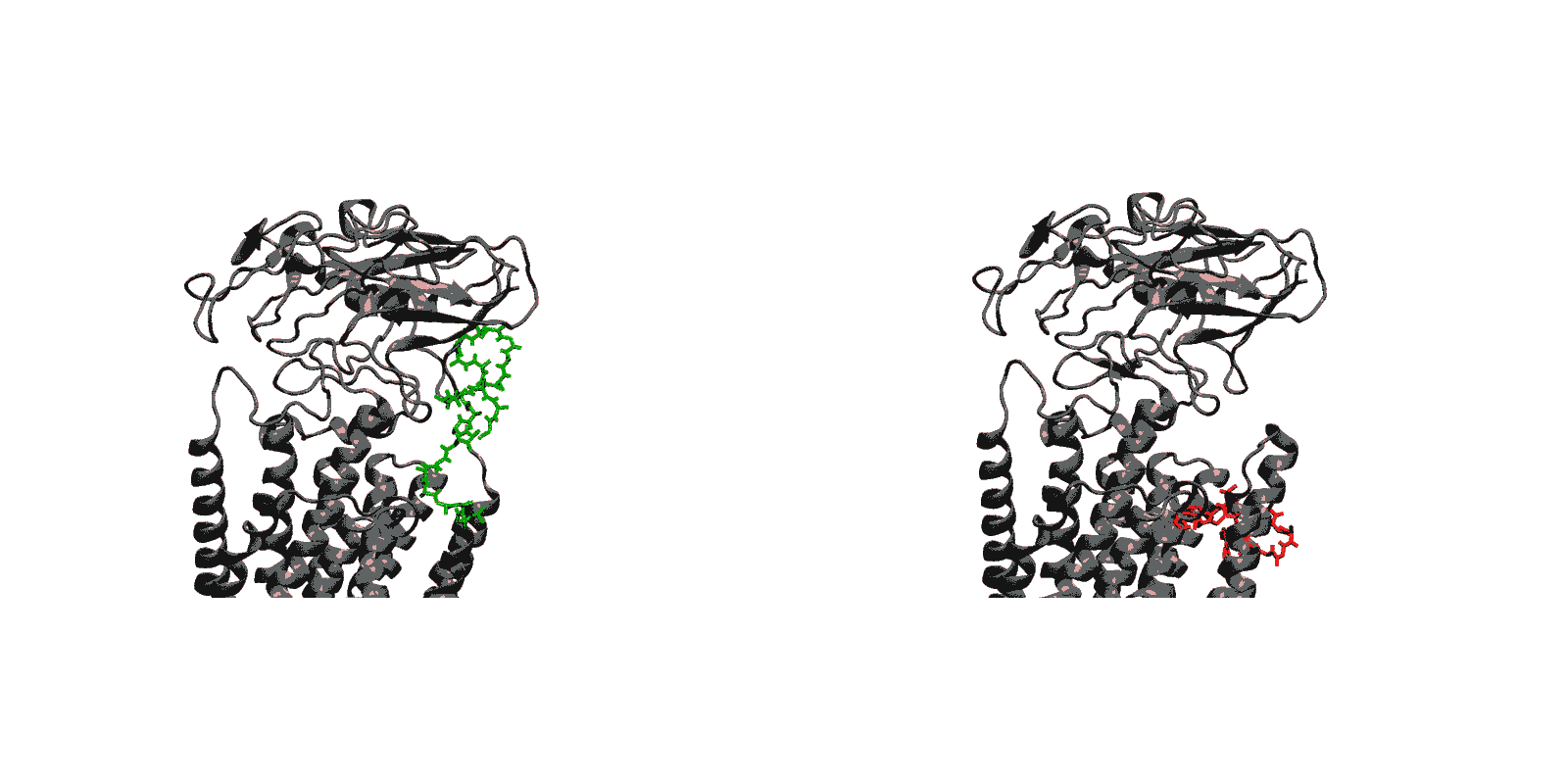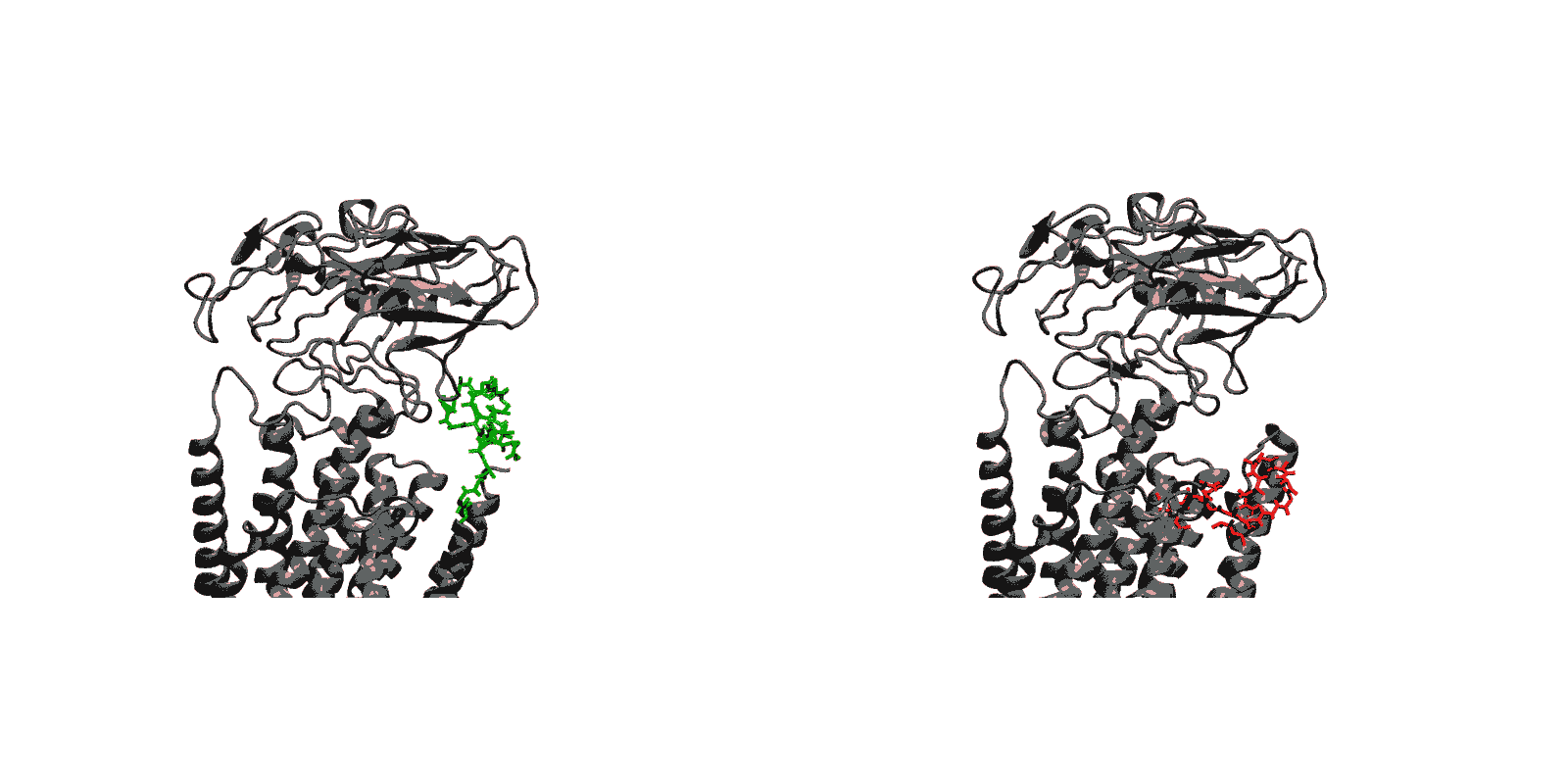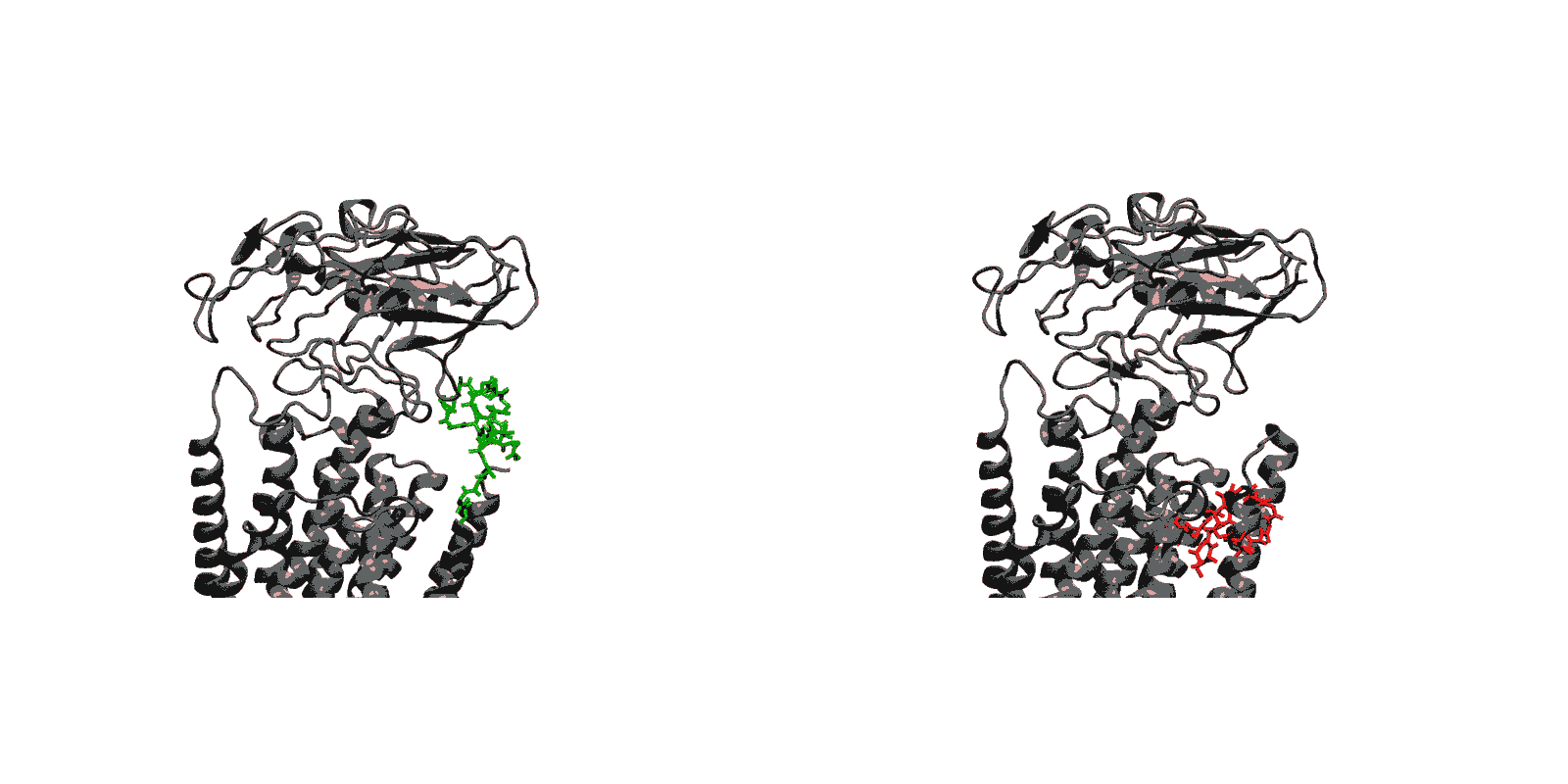
Background
In recent years, high-throughput genomic and proteomic projects have been revealing new genetic information at a very fast pace. The proper interpretation of such information in the context of the underlying biological mechanisms generally requires studies on a molecular level through biophysical and biochemical experiments as well as biomolecular modeling. Hence, the use of biophysical probes that can be easily integrated into protein structures for functional and structural characterization is becoming increasingly common. Lanthanide-binding tags (LBTs) are a class of such probes that are optimized for binding lanthanide ions.
We have created a fully automated webserver for the prediction of LBT insertions onto existing protein structures. The underlying method uses a coarse-grained backbone+Cβ representation of the protein and samples on the backbone dihedral angles. The method predicts the proper conformation of the LBT tag with respect to the parent protein in existing fusion crystal structures. On multiple membrane protein systems, the method’s prediction of feasibility of LBT insertion on various sites qualitatively agree with the experimental data. The method currently reports several metrics to assess the quality of LBT insertion in a specific site in a parent protein. We expect the method to serve as a useful computational tool for experimentalists by helping them select reasonable sites in a given parent protein where the LBT insertion will likely be successful.
Goals
- Given a parent protein where the LBT tag is to be inserted, the method will predict the likelihood of a successful LBT insertion at a certain site in the parent protein.
- Given a parent protein and a site of insertion, the method will predict the proper conformation of the parent+LBT fusion conformation.
Webserver
This webserver was developed by Aashish Adhikari from Tobin Sosnick’s group at the University of Chicago. Please contact us with questions or suggestions using the comments form below.







