Automated Force Field Parameterization for Nonpolarizable and Polarizable Atomic Models Based on Ab Initio Target Data
By Lei Huang and Benoı̂t Roux.
Published in Journal of Chemical Theory and Computation [Epub ahead of print] on July 12, 2013;9(8). PMID: 24223528. PMCID:3819940. Link to publication page
Core Facility: Computational Modeling

Abstract
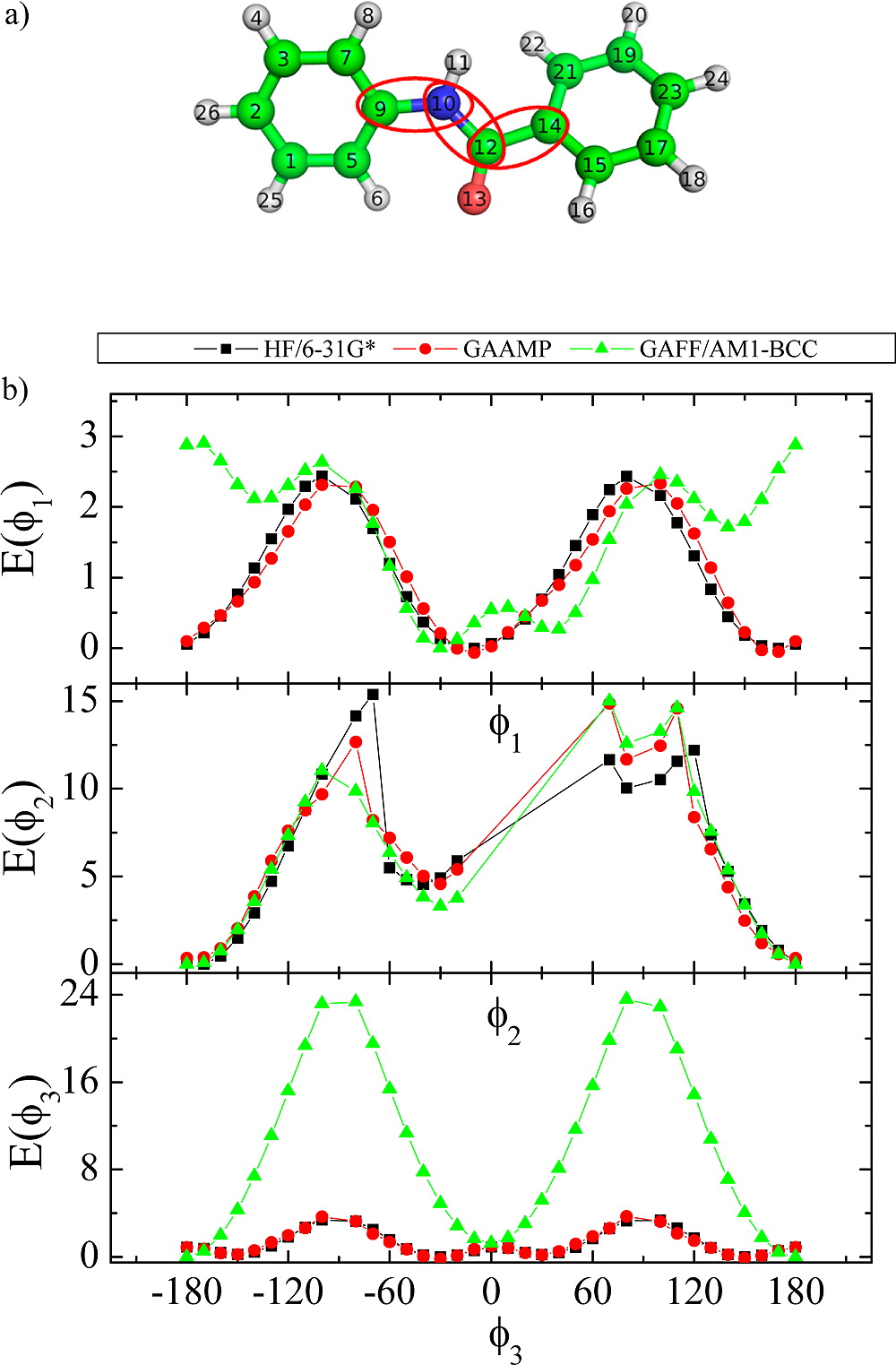
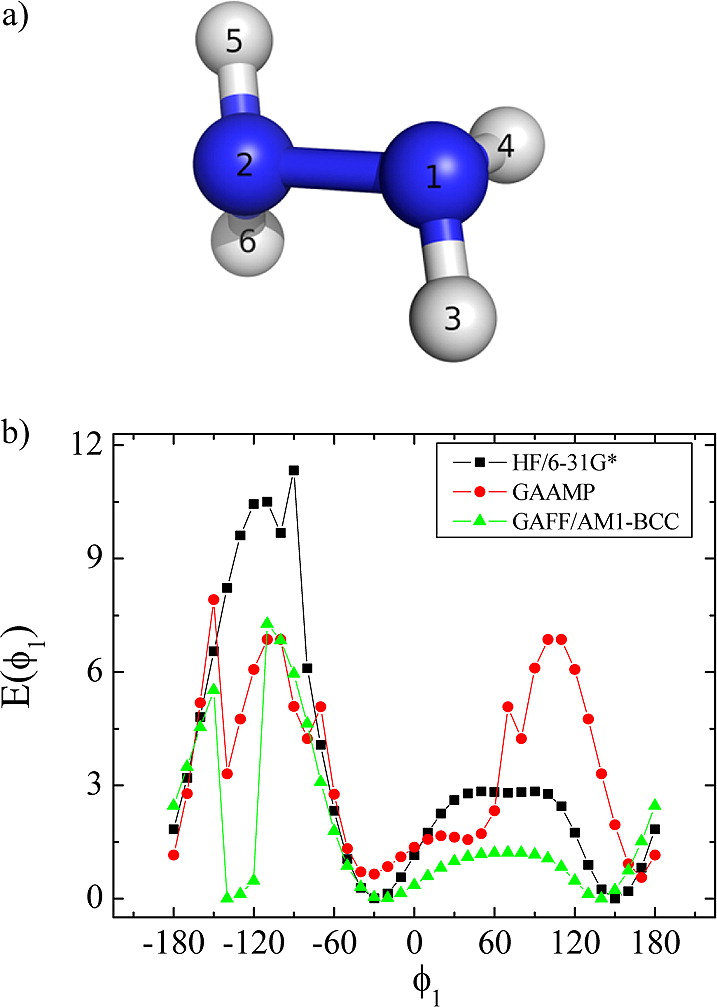
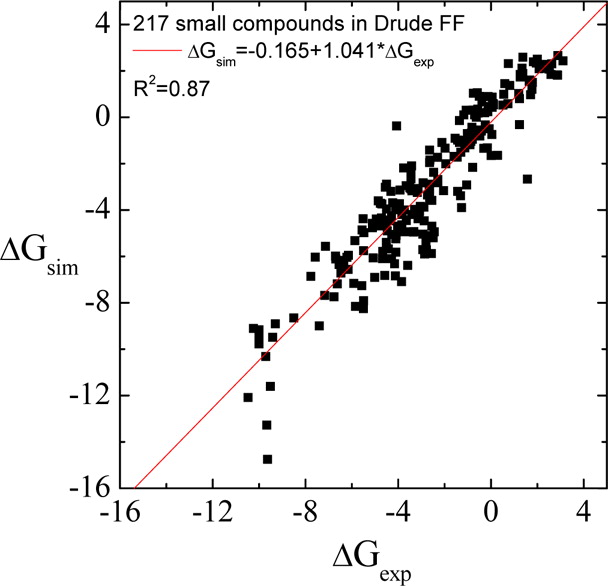
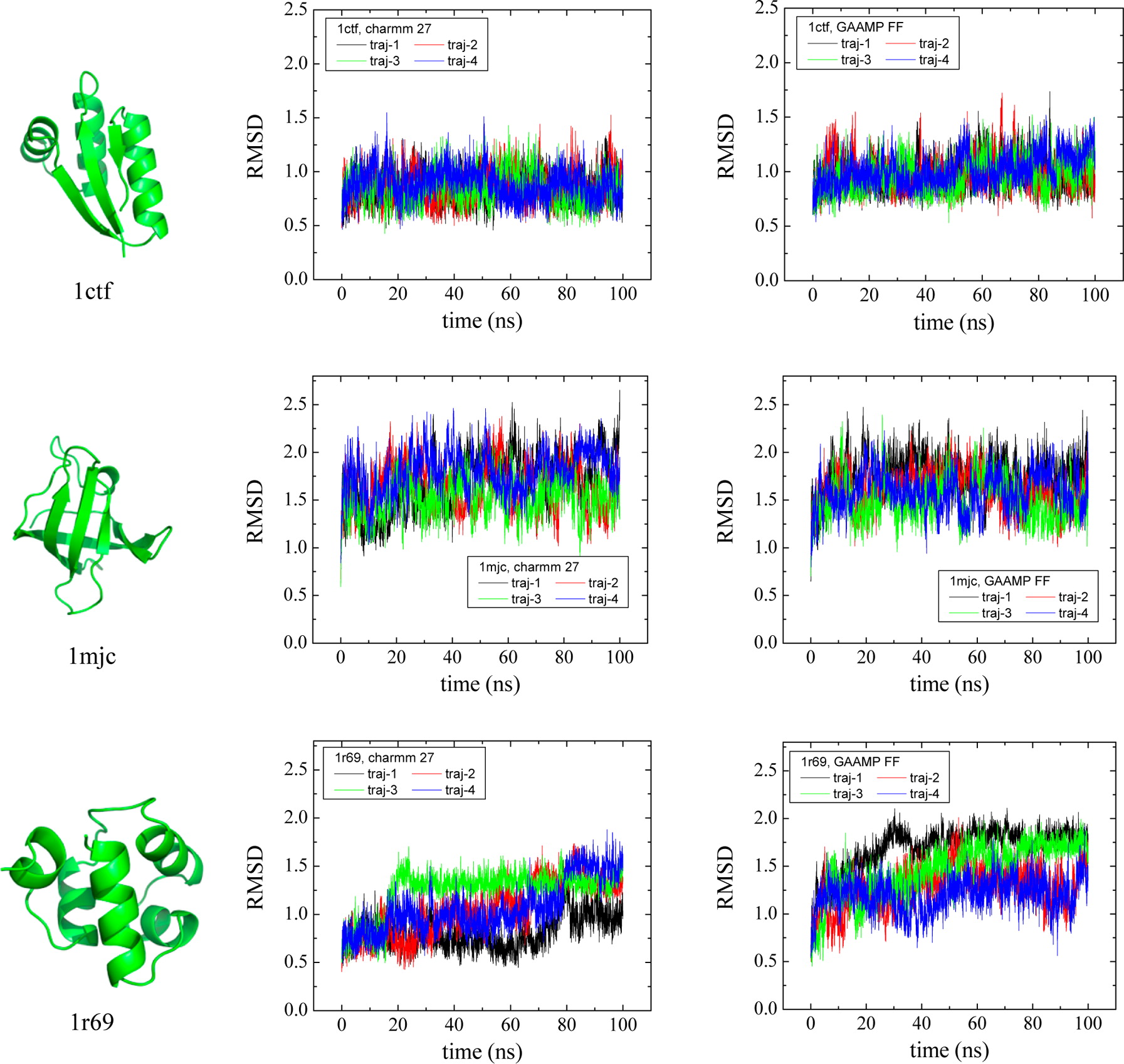
Classical molecular dynamics (MD) simulations based on atomistic models are increasingly used to study a wide range of biological systems. A prerequisite for meaningful results from such simulations is an accurate molecular mechanical force field. Most biomolecular simulations are currently based on the widely used AMBER and CHARMM force fields, which were parametrized and optimized to cover a small set of basic compounds corresponding to the natural amino acids and nucleic acid bases. Atomic models of additional compounds are commonly generated by analogy to the parameter set of a given force field. While this procedure yields models that are internally consistent, the accuracy of the resulting models can be limited. In this work, we propose a method, general automated atomic model parameterization (GAAMP), for generating automatically the parameters of atomic models of small molecules using the results from ab initio quantum mechanical (QM) calculations as target data. Force fields that were previously developed for a wide range of model compounds serve as initial guesses, although any of the final parameter can be optimized. The electrostatic parameters (partial charges, polarizabilities, and shielding) are optimized on the basis of QM electrostatic potential (ESP) and, if applicable, the interaction energies between the compound and water molecules. The soft dihedrals are automatically identified and parametrized by targeting QM dihedral scans as well as the energies of stable conformers. To validate the approach, the solvation free energy is calculated for more than 200 small molecules and MD simulations of three different proteins are carried out.