Constant electric field simulations of the membrane potential illustrated with simple systems
By James Gumbart, Fatemeh Khalili-Araghib, Marcos Sotomayor, and Benoît Roux.
Published in Biochimica et Biophysica Acta Feb 2012;1818(2):294-302. PMID: 22001851. PMCID: PMC3575077. Link to Pubmed page.
Core Facility: Computational Modeling
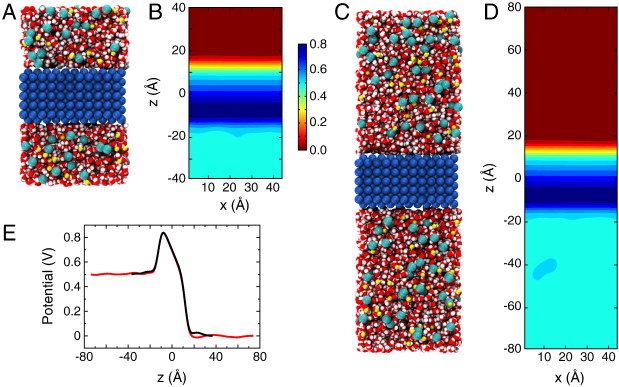
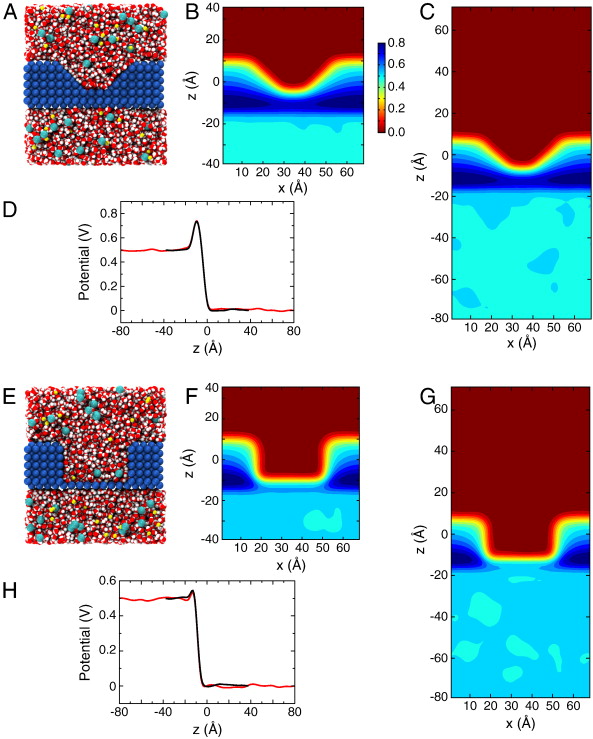
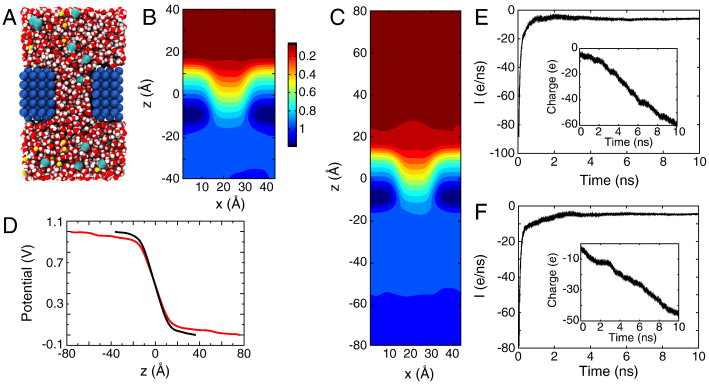
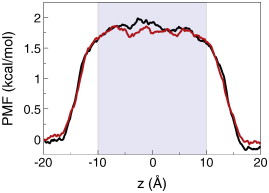
Figure 2. Membrane slab systems. (A) Full simulation system for the simple membrane slab shown as space-filling spheres with the membrane colored in blue; the water in red and white; and Na+ and Cl− ions in yellow and cyan, respectively. (B) Time-averaged electrostatic potential for the system in (A). The color gradient to the right indicates the scale for the potential in units of Volts. (C, D) Simulation system and potential identical to those in (A,B) except with the system length doubled and the applied field halved. (E) Potential along the z-axis for the system in A (black) and in C (red).
Abstract
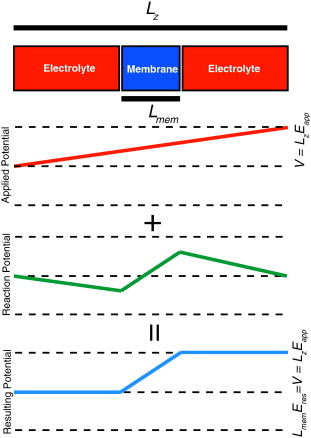
Advances in modern computational methods and technology make it possible to carry out extensive molecular dynamics simulations of complex membrane proteins based on detailed atomic models. The ultimate goal of such detailed simulations is to produce trajectories in which the behavior of the system is as realistic as possible. A critical aspect that requires consideration in the case of biological membrane systems is the existence of a net electric potential difference across the membrane. For meaningful computations, it is important to have well validated methodologies for incorporating the latter in molecular dynamics simulations. A widely used treatment of the membrane potential in molecular dynamics consists of applying an external uniform electric field E perpendicular to the membrane. The field acts on all charged particles throughout the simulated system, and the resulting applied membrane potential V is equal to the applied electric field times the length of the periodic cell in the direction perpendicular to the membrane. A series of test simulations based on simple membrane-slab models are carried out to clarify the consequences of the applied field. These illustrative tests demonstrate that the constant-field method is a simple and valid approach for accounting for the membrane potential in molecular dynamics studies of biomolecular systems. This article is part of a Special Issue entitled: Membrane protein structure and function.